Introduction
import requests
token = "your token here";
endpoint = "any valid endpoint to the service";
data = { }; #json with your input
#for POST methods
requests.post(endpoint, headers={"Authorization" : token}, json = data);
#for GET methods
requests.get(endpoint, headers={"Authorization" : token});
Clinical laboratory results are ubiquitous in any diagnosis making. Predicting abnormal values of not prescribed tests based on the results of performed tests looks intriguing, as it would be possible to make early diagnosis available to everyone. The special place is taken by the Common Blood Count (CBC) test, as it is the most widely used clinical procedure. Combining routine biochemical panels with CBC presents a set of test-value pairs that varies from patient to patient, or, in common settings, a table with missing values.
In our article published by IEEE, we formulate a tabular modeling problem as a set translation problem where the source set comprises pairs of GPT-like label column embedding and its corresponding value while the target set consists of the same type embeddings only. The proposed approach can effectively deal with missing values without implicitly estimating them and bridges the world of LLM with the tabular domain. Applying this method to clinical laboratory data, we achieve an improvement up to 8% AUC for joint predictions of high uric acid, glucose, cholesterol, and low ferritin levels, preliminary arXiv version. Russian version is also available
To cite our work please use the BibTex entry below:
This services provides API to get prediction based on our ULM-like models.
Authorization
import requests
data = {
"email": "valid email here",
"affiliation": "valid affiliation here",
"name": "valid company name here"
}
requests.post("https://ulm.roslis.ru/api/v1/register", json = data)
The service uses Bearer authentification, i.e. every request to the server should have a header "Authorization" with your valid token. To obtain a valid Bearer token, you need to use the company registration request. The required data includes the affiliation, email, and fullname.
After the data is verified, the token will be sent to the specified email address. As this operation is manual, 1-2 working days may need to accept your request.
You can request an access token by the authorization request or directly by email to ulm at roslis.ru. In the request please, submit your fullname, affiliation and your plans for using the service.
Input parameters
import requests
import json
token = "your authorization token"
def predict():
#unique identifier in your system, it may be an artificial ID or a barcode.
sample = "0012"
# submitt all available data for results of tests shown in the table
# key is a test LOINC or its name (age, gender) and the value is corresponding result
params = {"GENDER" : "1", "AGE" : "45", "804-5" : "4.5900",
"789-8" : "4.74", "30350-3" : "139.0",
"71829-6" : "86", "777-3" : "243",
"768-2" : "52", "711-2" : "4",
"704-7" : "1", "737-7": "33", "5905-5" : "10"}
#This example reads as "Sample 0012 of a male 45 years old has
# WBC = 4.59, RBC = 4.74, HGB = 139, MCV = 86, PLT = 243,
# NEUT = 52, EOS = 4, BASO = 1, LYMPH = 33, MONO = 10".
data = [ {"sample" : sample, "results" : params } ]
ret = requests.post("https://ulm.roslis.ru/api/v1/models/1",
headers={"Authorization" : token}, json = data)
res = json.loads(ret.content)
print(json.dumps(res, ensure_ascii=False))
predict()
The above command returns JSON structured like this in case of success:
{
"data": [
{
"results": [
{
"analyte": "Prognosis of low ferritin level",
"conclusion": "negative",
"result": "0.004286268"
}
],
"sample": "0012"
}
],
"status": "ok"
}
or in case of wrong input (out of applicability domain):
{
"data": [
{
"results": [
{
"analyte": "Prognosis of low ferritin level",
"conclusion": "out of applicability domain",
"result": "n/a"
}
],
"sample": "0012"
}
],
"status": "ok"
}
or technical failure:
{
"data": [
{ }
],
"comments" : "The server is unavailable, please try again later.",
"status": "failed"
}
The ULM services provides a number of models that differ in input and output parameters and the underlying architecture as well. For the full list of models and their statistics reffer to the method models. If it is not stressed explicitly, parameters for the models are a subset from:
| Parameter (Analyte) | LOINC | Acceptable Ranges | Units | Obligatory |
|---|---|---|---|---|
| Age | - | 1 - 110 | Yeras | Y |
| Alanine transaminase, (ALT) | 1742-6 | 0.10 - 1721.40 | U/L | N |
| Albumin, (ALB) | 1751- 7 | 0.04 - 60.14 | g/L | N |
| Aspartate aminotransferase, (AST) | 1920-8 | 0.30 - 1518.50 | U/L | N |
| Basophils, (BASO) | 704-7 | 0.02 - 27.20 | % | P |
| Cholesterol, (CHOL) | 14647-2 | 0.02 - 17.20 | mmol/L | N |
| Creatinine, (CREA) | 14682-9 | 0.30 - 1618.80 | μmol/L | N |
| C-reactive protein, (CRP) | 1988-5 | 0.01 - 250.95 | mg/L | N |
| Direct bilirubin, (BC) | 29760-6 | 0.04 - 238.40 | μmol/L | N |
| Eosinophils, (EOS) | 711-2 | 0.09 - 43.80 | % | P |
| Ferritin, (FER) | 20567-4 | 0.01 - 1667.20 | μg/L | N |
| Folic acid, (FOL) | 2284-8 | 0.56 - 330.11 | ng/mL | N |
| Gender, (Male =1, Female = 0) | - | 0 or 1 | - | Y |
| Glucose, (GLU) | 14771-0 | 0.01 - 26.77 | mmol/L | N |
| Glycated hemoglobin | 4548-4 | 2.2 - 21.5 | % | N |
| Granulocytes, (GRA) | 19023-1 | 14.7 - 94.7 | % | P |
| Hemoglobin, (HGB) | 30350-3 | 11.00 - 215.00 | g/L | Y |
| Indirect bilirubin, (BU) | 14630-8 | 0.05 - 220.79 | μmol/L | N |
| Lactate dehydrogenase, (LDH) | 2532-0 | 2.00 - 4983.00 | U/L | N |
| Lymphocytes, (LYMPH) | 737-7 | 0.10 - 90.70 | % | P |
| Mean corpuscular volume, (MCV) | 71829-6 | 0.70 - 134.00 | fL | Y |
| Middle-size Cells, (MID) | 32155-4 | 1.2 - 29.4 | % | P |
| Monocytes, (MONO) | 5905-5 | 0.10 - 55.40 | % | P |
| Neutrophils, (NEUT) | 768-2 | 2.90 - 100.00 | % | P |
| Platelets, (PLT) | 777-3 | 1.00 - 1053.00 | 109/L | Y |
| Red blood cells, (RBC) | 789-8 | 0.24 - 8.21 | 1012/L | Y |
| Total bilirubin, (TBIL) | 54363-7 | 0.03 - 432.43 | μmol/L | N |
| Total protein, (PRO) | 13980-8 | 19.20 - 132.10 | g/L | N |
| Total PSA, (PSA) | 2857-1 | 0 - 4212 | ng/mL | N |
| Urea, (UREA) | 22664-7 | 0.50 - 67.50 | mmol/L | N |
| Uric acid, (URIC) | 14933-6 | 0.00 - 1.22 | mmol/L | N |
| Vitamin B12, (VB) | 2132-9 | 1.00 - 39833.00 | pg/mL | Y |
| White blood cells, (WBC) | 804-5 | 0.10 - 68.70 | 109/L | Y |
As a rule of thumb, every model requires basic CBC data, other parameters are optional. If the CBC data has 5-DIFF, then NEUT, MONO, EOS, BASO, and LYMP has to be supplied, in case of 3-DIFF - MID, GRA, and LYMP (these analytes are marked as P(artial) in the Table, indicating at least one group of parameters 5-DIFF or 3-DIFF should be provided.
The input to any model is a json array, each item contains the request identificator and a variable length map of pairs (LOINC, result) as shown in the example.
Available models
import requests
import json
token = "your authorization token"
def showModels():
ret = requests.get("https://ulm.roslis.ru/api/v1/models",
headers={"Authorization" : token});
res = json.loads(ret.content);
print(json.dumps(res, ensure_ascii=False));
showModels();
As all the models solve classification problems, we provide Accuracy, Sensitivity, Specificity, F1, and DOR (diagnostic odds ratio). All the values are calculated for the test set with a threshold calibrated on train dataset.
Applying the model might require recalibration. You have to do it by yourself as the service does not provide such functionality.
To get a list of all available models, use
GET https://ulm.roslis.ru/api/v1/models
Basic ferritin model
This model has internal index 1 and make a prognosis of low ferritin levels, < 12 μg/L (Some laboratories may use ng/mL as units, ng/mL = μg/L). It is a shallow network calculating a probability of low levels of ferritin for provided data. The statistics of the model is:
| AUC,% | N | Accuracy, % | Sensitivity, % | Specificity, % | F1 | DOR | Threshold |
|---|---|---|---|---|---|---|---|
| 88.75 | 106065 | 80.46 | 81.48 | 80.33 | 0.49 | 18 | 0.10667945 |
HTTP Requst
POST https://ulm.roslis.ru/api/v1/models/1
Parameters
CBC data is obligatory, other tests may be supplied and therefore used as well.
Vanilla ULM model
import requests
import json
token = "your authorization token"
def predict():
sample = "0013"; #unique identifier in your system
params = {"GENDER" : "0", "AGE" : "24", "804-5" : "4.59",
"789-8" : "4.74", "30350-3" : "152", "71829-6" : "86",
"777-3" : "243", "768-2" : "52", "711-2" : "4",
"704-7" : "1", "737-7": "33", "5905-5" : "10"};
data = [ {"sample" : sample, "results" : params } ];
ret = requests.post("https://ulm.roslis.ru/api/v1/models/2",
headers={"Authorization" : token}, json = data);
res = json.loads(ret.content);
print(json.dumps(res, ensure_ascii=False));
predict();
The above command returns JSON structured like this:
{
"data": [
{
"results": [
{
"analyte": "Prognosis of low ferritin level",
"conclusion": "negative",
"result": "0.0246749"
},
{
"analyte": "Prognosis of high glucose level",
"conclusion": "negative",
"result": "0.004755818"
},
{
"analyte": "Prognosis of high cholesterol level",
"conclusion": "positive",
"result": "0.29461566"
},
{
"analyte": "Prognosis of high uric acid level",
"conclusion": "negative",
"result": "0.12722556"
}
],
"sample": "0013"
}
],
"status": "ok"
}
Our first vanilla ULM model that is described in our paper. The model jointly predicts low ferritin level, and high levels for glucose, cholesterol, and uric acid.
| Parameter | AUC,% | N | Accuracy, % | Sensitivity, % | Specificity, % | F1 | DOR | Threshold |
|---|---|---|---|---|---|---|---|---|
| Ferritin | 89.7 | 105928 | 80.9 | 79.9 | 80.9 | 0.48 | 16.6 | 0.13470852 |
| Glucose | 82.4 | 680502 | 74.5 | 73.3 | 74.6 | 0.23 | 7.9 | 0.06627314 |
| Cholesterol | 77.9 | 248806 | 70.1 | 70.1 | 70.1 | 0.73 | 5.6 | 0.61889935 |
| Uric acid | 79.7 | 92901 | 70.5 | 68.3 | 70.9 | 0.40 | 5.3 | 0.18092427 |
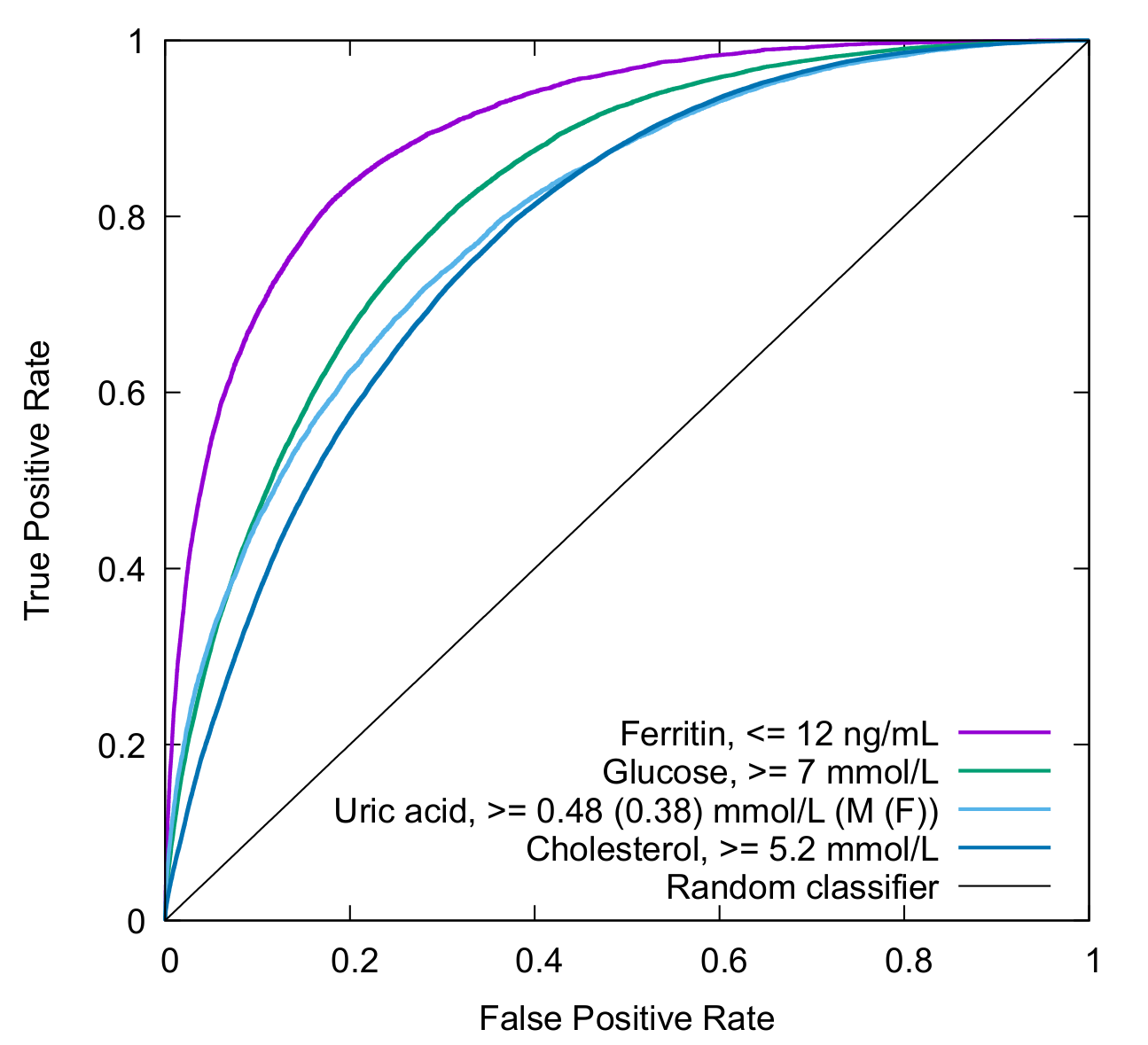
HTTP Requst
POST https://ulm.roslis.ru/api/v1/models/2
Parameters
CBC data is obligatory, other tests may be supplied and therefore used as well.
ULM model (FER-URIC-HbA1c)
| Parameter | AUC,% | N | Accuracy, % | Sensitivity, % | Specificity, % | F1 | DOR | Threshold |
|---|---|---|---|---|---|---|---|---|
| Ferritin | 89.4 | 105928 | 80.8 | 80.5 | 80.8 | 0.48 | 17.4 | 0.10442664 |
| Uric acid | 80.2 | 92901 | 71.4 | 73.4 | 74.4 | 0.34 | 7.6 | 0.10641282 |
| HbA1c | 90.1 | 37925 | 80.2 | 79.5 | 80.5 | 0.71 | 16.0 | 0.32560816 |
HTTP Requst
POST https://ulm.roslis.ru/api/v1/models/3
Errors
Known issues

